LETID - Outdoor Environments#
This is an example on how to model LETID progression in outdoor environments
We can use the equations in this library to model LETID progression in a simulated outdoor environment, given that we have weather and system data. This example makes use of tools from the fabulous pvlib library to calculate system irradiance and temperature, which we use to calculate progression in LETID states.
This will illustrate the potential of “Temporary Recovery”, i.e., the backwards transition of the LETID defect B->A that can take place with carrier injection at lower temperatures.
Requirements:
pvlib,pandas,numpy,matplotlib
Objectives:
Use
pvliband provided weather files to set up a temperature and injection timeseriesDefine necessary solar cell device parameters
Define necessary degradation parameters: degraded lifetime and defect states
Run through timeseries, calculating defect states
Calculate device degradation and plot
# if running on google colab, uncomment the next line and execute this cell to install the dependencies and prevent "ModuleNotFoundError" in later cells:
# !pip install pvdeg==0.3.3
from pvdeg import letid, collection, utilities, DATA_DIR
import pvlib
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import pvdeg
# This information helps with debugging and getting support :)
import sys
import platform
print("Working on a ", platform.system(), platform.release())
print("Python version ", sys.version)
print("Pandas version ", pd.__version__)
print("pvlib version ", pvlib.__version__)
print("pvdeg version ", pvdeg.__version__)
Working on a Linux 6.11.0-1018-azure
Python version 3.11.13 (main, Jun 4 2025, 04:12:12) [GCC 13.3.0]
Pandas version 2.3.3
pvlib version 0.13.1
pvdeg version 0.1.dev1+g51172fdc5
First, we’ll use pvlib to create and run a model system, and use the irradiance, temperature, and operating point of that model to set up our LETID model For this example, we’ll model a fixed latitude tilt system at NREL, in Golden, CO, USA, using NSRDB hourly PSM weather data, SAPM temperature models, and module and inverter models from the CEC database.
# load weather and location data, use pvlib read_psm3 function with map_variables = True
sam_file = "psm3.csv"
weather, meta = pvdeg.weather.read(
os.path.join(DATA_DIR, sam_file), file_type="PSM3", map_variables=True
)
weather
| Year | Month | Day | Hour | Minute | dni | dhi | ghi | temp_air | dew_point | wind_speed | relative_humidity | poa_global | temp_cell | temp_module | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1999-01-01 00:30:00-07:00 | 1999 | 1 | 1 | 0 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -5.0 | 1.8 | 79.39 | 0.0 | 0.0 | 0.0 |
| 1999-01-01 01:30:00-07:00 | 1999 | 1 | 1 | 1 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.7 | 80.84 | 0.0 | 0.0 | 0.0 |
| 1999-01-01 02:30:00-07:00 | 1999 | 1 | 1 | 2 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.5 | 82.98 | 0.0 | 0.0 | 0.0 |
| 1999-01-01 03:30:00-07:00 | 1999 | 1 | 1 | 3 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.3 | 85.01 | 0.0 | 0.0 | 0.0 |
| 1999-01-01 04:30:00-07:00 | 1999 | 1 | 1 | 4 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.3 | 85.81 | 0.0 | 0.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1999-12-31 19:30:00-07:00 | 1999 | 12 | 31 | 19 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -3.0 | 0.9 | 83.63 | 0.0 | 0.0 | 0.0 |
| 1999-12-31 20:30:00-07:00 | 1999 | 12 | 31 | 20 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -3.0 | 1.2 | 86.82 | 0.0 | 0.0 | 0.0 |
| 1999-12-31 21:30:00-07:00 | 1999 | 12 | 31 | 21 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.6 | 83.78 | 0.0 | 0.0 | 0.0 |
| 1999-12-31 22:30:00-07:00 | 1999 | 12 | 31 | 22 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -4.0 | 1.7 | 81.22 | 0.0 | 0.0 | 0.0 |
| 1999-12-31 23:30:00-07:00 | 1999 | 12 | 31 | 23 | 30 | 0.0 | 0.0 | 0.0 | 0.0 | -5.0 | 1.8 | 79.43 | 0.0 | 0.0 | 0.0 |
8760 rows × 15 columns
# if our weather file doesn't have precipitable water, calculate it with pvlib
if "precipitable_water" not in weather.columns:
weather["precipitable_water"] = pvlib.atmosphere.gueymard94_pw(
weather["temp_air"], weather["relative_humidity"]
)
# rename some columns for pvlib if they haven't been already
weather.rename(
columns={
"GHI": "ghi",
"DNI": "dni",
"DHI": "dhi",
"Temperature": "temp_air",
"Wind Speed": "wind_speed",
"Relative Humidity": "relative_humidity",
"Precipitable Water": "precipitable_water",
},
inplace=True,
)
weather = weather[
[
"ghi",
"dni",
"dhi",
"temp_air",
"wind_speed",
"relative_humidity",
"precipitable_water",
]
]
weather
| ghi | dni | dhi | temp_air | wind_speed | relative_humidity | precipitable_water | |
|---|---|---|---|---|---|---|---|
| 1999-01-01 00:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.8 | 79.39 | 0.891869 |
| 1999-01-01 01:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 | 80.84 | 0.908158 |
| 1999-01-01 02:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 82.98 | 0.932199 |
| 1999-01-01 03:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 85.01 | 0.955004 |
| 1999-01-01 04:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 85.81 | 0.963991 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1999-12-31 19:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | 83.63 | 0.939501 |
| 1999-12-31 20:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 86.82 | 0.975338 |
| 1999-12-31 21:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.6 | 83.78 | 0.941186 |
| 1999-12-31 22:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 | 81.22 | 0.912427 |
| 1999-12-31 23:30:00-07:00 | 0.0 | 0.0 | 0.0 | 0.0 | 1.8 | 79.43 | 0.892318 |
8760 rows × 7 columns
# import pvlib stuff and pick a module and inverter. Choice of these things will slightly affect the pvlib results which we later use to calculate injection.
# we'll use the SAPM temperature model open-rack glass/polymer coeffecients.
from pvlib.temperature import TEMPERATURE_MODEL_PARAMETERS
from pvlib.location import Location
from pvlib.pvsystem import PVSystem
from pvlib.modelchain import ModelChain
cec_modules = pvlib.pvsystem.retrieve_sam("CECMod")
cec_inverters = pvlib.pvsystem.retrieve_sam("cecinverter")
cec_module = cec_modules["Jinko_Solar_Co___Ltd_JKM260P_60"]
cec_inverter = cec_inverters["ABB__ULTRA_750_TL_OUTD_1_US_690_x_y_z__690V_"]
temperature_model_parameters = TEMPERATURE_MODEL_PARAMETERS["sapm"][
"open_rack_glass_polymer"
]
# set up system in pvlib
lat = meta["latitude"]
lon = meta["longitude"]
tz = meta["tz"]
elevation = meta["altitude"]
surface_tilt = lat # fixed, latitude tilt
surface_azimuth = 180 # south-facing
location = Location(lat, lon, tz, elevation, "Golden, CO, USA")
system = PVSystem(
surface_tilt=surface_tilt,
surface_azimuth=surface_azimuth,
module_parameters=cec_module,
inverter_parameters=cec_inverter,
temperature_model_parameters=temperature_model_parameters,
)
# create and run pvlib modelchain
mc = ModelChain(system, location, aoi_model="physical")
mc.run_model(weather)
ModelChain:
name: None
clearsky_model: ineichen
transposition_model: haydavies
solar_position_method: nrel_numpy
airmass_model: kastenyoung1989
dc_model: cec
ac_model: sandia_inverter
aoi_model: physical_aoi_loss
spectral_model: no_spectral_loss
temperature_model: sapm_temp
losses_model: no_extra_losses
Set up timeseries#
In this example, injection is a function of both the operating point of the module (which we will assume is maximum power point) and irradiance. Maximum power point injection is equivalent to \((I_{sc}-I_{mp})/I_{sc}\times Ee\), where \(Ee\) is effective irradiance, the irradiance absorbed by the module’s cells. We normalize it to 1-sun irradiance, 1000 \(W/m^2\).
We will use the irradiance, DC operating point, and cell temperature from the pvlib modelchain results.
ee = mc.results.effective_irradiance
# injection = (mc.results.dc['i_sc']-mc.results.dc['i_mp'])/(mc.results.dc['i_sc'])*(ee/1000)
injection = letid.calc_injection_outdoors(mc.results)
temperature = mc.results.cell_temperature
timesteps = pd.DataFrame(
{"Temperature": temperature, "Injection": injection}
) # create a DataFrame with cell temperature and injection
timesteps.reset_index(
inplace=True
) # reset the index so datetime is a column. I prefer integer indexing.
timesteps.rename(columns={"index": "Datetime"}, inplace=True)
# filter out times when injection is NaN, these won't progress LETID, and it'll make the calculations below run faster
timesteps = timesteps[timesteps["Injection"].notnull()]
timesteps.reset_index(inplace=True, drop=True)
timesteps
| Datetime | Temperature | Injection | |
|---|---|---|---|
| 0 | 1999-01-01 08:30:00-07:00 | 3.348774 | 0.006094 |
| 1 | 1999-01-01 09:30:00-07:00 | 12.378439 | 0.031193 |
| 2 | 1999-01-01 10:30:00-07:00 | 16.595081 | 0.044058 |
| 3 | 1999-01-01 11:30:00-07:00 | 17.457240 | 0.043239 |
| 4 | 1999-01-01 12:30:00-07:00 | 6.392783 | 0.007145 |
| ... | ... | ... | ... |
| 4296 | 1999-12-31 12:30:00-07:00 | 9.515577 | 0.001159 |
| 4297 | 1999-12-31 13:30:00-07:00 | 28.119796 | 0.044562 |
| 4298 | 1999-12-31 14:30:00-07:00 | 23.314672 | 0.034774 |
| 4299 | 1999-12-31 15:30:00-07:00 | 17.890528 | 0.027285 |
| 4300 | 1999-12-31 16:30:00-07:00 | 4.552365 | 0.001190 |
4301 rows × 3 columns
Device parameters#
To define a device, we need to define several important quantities about the device: wafer thickness (in \(\mu m\)), rear surface recombination velocity (in cm/s), and cell area (in cm2).
wafer_thickness = 180 # um
s_rear = 46 # cm/s
cell_area = 243 # cm^2
Other device parameters Other required device parameters: base diffusivity (in cm2/s), and optical generation profile, which allow us to estimate current collection in the device.
generation_df = pd.read_excel(
os.path.join(DATA_DIR, "PVL_GenProfile.xlsx"), header=0
) # this is an optical generation profile generated by PVLighthouse's OPAL2 default model for 1-sun, normal incident AM1.5 sunlight on a 180-um thick SiNx-coated, pyramid-textured wafer.
generation = generation_df["Generation (cm-3s-1)"]
depth = generation_df["Depth (um)"]
d_base = 27 # cm^2/s electron diffusivity. See https://www2.pvlighthouse.com.au/calculators/mobility%20calculator/mobility%20calculator.aspx for details
Degradation parameters#
To model the device’s degradation, we need to define several more important quantities about the degradation the device will experience. These include undegraded and degraded lifetime (in \(\mu s\)).
tau_0 = 115 # us, carrier lifetime in non-degraded states, e.g. LETID/LID states A or C
tau_deg = 55 # us, carrier lifetime in fully-degraded state, e.g. LETID/LID state B
Remaining degradation parameters:
The rest of the quantities to define are: the initial percentage of defects in each state (A, B, and C), and the dictionary of mechanism parameters.
In this example, we’ll assume the device starts in the fully-undegraded state (100% state A), and we’ll use the kinetic parameters for LETID degradation from Repins.
# starting defect state percentages
nA_0 = 100
nB_0 = 0
nC_0 = 0
mechanism_params = utilities.get_kinetics("repins")
timesteps[["NA", "NB", "NC", "tau"]] = (
np.nan
) # create columns for defect state percentages and lifetime, fill with NaNs for now, to fill iteratively below
timesteps.loc[0, ["NA", "NB", "NC"]] = (
nA_0,
nB_0,
nC_0,
) # assign first timestep defect state percentages
timesteps.loc[0, "tau"] = letid.tau_now(
tau_0, tau_deg, nB_0
) # calculate tau for the first timestep
Run through timesteps#
Since each timestep depends on the preceding timestep, we need to calculate in a loop. This will take a few minutes depending on the length of the timeseries.
for index, timestep in timesteps.iterrows():
# first row tau has already been assigned
if index == 0:
# calc device parameters for first row
tau = tau_0
jsc = collection.calculate_jsc_from_tau_cp(
tau, wafer_thickness, d_base, s_rear, generation, depth
)
voc = letid.calc_voc_from_tau(tau, wafer_thickness, s_rear, jsc, temperature=25)
timesteps.at[index, "Jsc"] = jsc
timesteps.at[index, "Voc"] = voc
# loop through rows, new tau calculated based on previous NB. Reaction proceeds based on new tau.
else:
n_A = timesteps.at[index - 1, "NA"]
n_B = timesteps.at[index - 1, "NB"]
n_C = timesteps.at[index - 1, "NC"]
tau = letid.tau_now(tau_0, tau_deg, n_B)
jsc = collection.calculate_jsc_from_tau_cp(
tau, wafer_thickness, d_base, s_rear, generation, depth
)
temperature = timesteps.at[index, "Temperature"]
injection = timesteps.at[index, "Injection"]
# calculate defect reaction kinetics: reaction constant and carrier concentration factor.
k_AB = letid.k_ij(
mechanism_params["v_ab"], mechanism_params["ea_ab"], temperature
)
k_BA = letid.k_ij(
mechanism_params["v_ba"], mechanism_params["ea_ba"], temperature
)
k_BC = letid.k_ij(
mechanism_params["v_bc"], mechanism_params["ea_bc"], temperature
)
k_CB = letid.k_ij(
mechanism_params["v_cb"], mechanism_params["ea_cb"], temperature
)
x_ab = letid.carrier_factor(
tau,
"ab",
temperature,
injection,
jsc,
wafer_thickness,
s_rear,
mechanism_params,
)
x_ba = letid.carrier_factor(
tau,
"ba",
temperature,
injection,
jsc,
wafer_thickness,
s_rear,
mechanism_params,
)
x_bc = letid.carrier_factor(
tau,
"bc",
temperature,
injection,
jsc,
wafer_thickness,
s_rear,
mechanism_params,
)
# calculate the instantaneous change in NA, NB, and NC
dN_Adt = (k_BA * n_B * x_ba) - (k_AB * n_A * x_ab)
dN_Bdt = (
(k_AB * n_A * x_ab) + (k_CB * n_C) - ((k_BA * x_ba + k_BC * x_bc) * n_B)
)
dN_Cdt = (k_BC * n_B * x_bc) - (k_CB * n_C)
t_step = (
timesteps.at[index, "Datetime"] - timesteps.at[index - 1, "Datetime"]
).total_seconds()
# assign new defect state percentages
timesteps.at[index, "NA"] = n_A + dN_Adt * t_step
timesteps.at[index, "NB"] = n_B + dN_Bdt * t_step
timesteps.at[index, "NC"] = n_C + dN_Cdt * t_step
# calculate device parameters
timesteps.at[index, "tau"] = tau
timesteps.at[index, "Jsc"] = jsc
timesteps.at[index, "Voc"] = letid.calc_voc_from_tau(
tau, wafer_thickness, s_rear, jsc, temperature=25
)
Finish calculating degraded device parameters.#
Now that we have calculated defect states, we can calculate all the quantities that depend on defect states.
timesteps["tau"] = letid.tau_now(tau_0, tau_deg, timesteps["NB"])
# calculate device Jsc for every timestep. Unfortunately this requires an integration so I think we have to run through a loop. Device Jsc allows calculation of device Voc.
for index, timestep in timesteps.iterrows():
jsc_now = collection.calculate_jsc_from_tau_cp(
timesteps.at[index, "tau"], wafer_thickness, d_base, s_rear, generation, depth
)
timesteps.at[index, "Jsc"] = jsc_now
timesteps.at[index, "Voc"] = letid.calc_voc_from_tau(
timesteps.at[index, "tau"], wafer_thickness, s_rear, jsc_now, temperature=25
)
timesteps = letid.calc_device_params(
timesteps, cell_area=243
) # this function quickly calculates the rest of the device parameters: Isc, FF, max power, and normalized max power
timesteps
| Datetime | Temperature | Injection | NA | NB | NC | tau | Jsc | Voc | Isc | FF | Pmp | Pmp_norm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1999-01-01 08:30:00-07:00 | 3.348774 | 0.006094 | 100.000000 | 0.000000 | 0.000000e+00 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| 1 | 1999-01-01 09:30:00-07:00 | 12.378439 | 0.031193 | 99.997579 | 0.002421 | 0.000000e+00 | 114.996963 | 41.590987 | 0.666327 | 10.106610 | 0.840987 | 5.663460 | 0.999999 |
| 2 | 1999-01-01 10:30:00-07:00 | 16.595081 | 0.044058 | 99.992005 | 0.007995 | 5.966711e-09 | 114.989971 | 41.590966 | 0.666326 | 10.106605 | 0.840986 | 5.663446 | 0.999996 |
| 3 | 1999-01-01 11:30:00-07:00 | 17.457240 | 0.043239 | 99.985976 | 0.014024 | 2.732911e-08 | 114.982409 | 41.590942 | 0.666324 | 10.106599 | 0.840986 | 5.663430 | 0.999994 |
| 4 | 1999-01-01 12:30:00-07:00 | 6.392783 | 0.007145 | 99.985711 | 0.014289 | 2.841966e-08 | 114.982076 | 41.590941 | 0.666324 | 10.106599 | 0.840986 | 5.663430 | 0.999993 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4296 | 1999-12-31 12:30:00-07:00 | 9.515577 | 0.001159 | 41.396088 | 54.267061 | 4.336851e+00 | 72.235986 | 41.384030 | 0.656856 | 10.056319 | 0.839304 | 5.544072 | 0.978918 |
| 4297 | 1999-12-31 13:30:00-07:00 | 28.119796 | 0.044562 | 41.396882 | 54.265942 | 4.337177e+00 | 72.236540 | 41.384034 | 0.656856 | 10.056320 | 0.839304 | 5.544074 | 0.978919 |
| 4298 | 1999-12-31 14:30:00-07:00 | 23.314672 | 0.034774 | 41.402923 | 54.259760 | 4.337317e+00 | 72.239600 | 41.384057 | 0.656857 | 10.056326 | 0.839305 | 5.544086 | 0.978921 |
| 4299 | 1999-12-31 15:30:00-07:00 | 17.890528 | 0.027285 | 41.415085 | 54.247542 | 4.337373e+00 | 72.245649 | 41.384102 | 0.656859 | 10.056337 | 0.839305 | 5.544109 | 0.978925 |
| 4300 | 1999-12-31 16:30:00-07:00 | 4.552365 | 0.001190 | 41.415660 | 54.246967 | 4.337373e+00 | 72.245934 | 41.384105 | 0.656859 | 10.056337 | 0.839305 | 5.544110 | 0.978925 |
4301 rows × 13 columns
Note of course that all these calculated device parameters are modeled STC device parameters, not the instantaneous, weather-dependent values. This isn’t a robust performance model of a degraded module.
Plot the results#
from cycler import cycler
plt.style.use("default")
fig, ax = plt.subplots()
ax.set_prop_cycle(
cycler("color", ["tab:blue", "tab:orange", "tab:green"])
+ cycler("linestyle", ["-", "--", "-."])
)
ax.plot(timesteps["Datetime"], timesteps[["NA", "NB", "NC"]].values)
ax.legend(labels=["$N_A$", "$N_B$", "$N_C$"], loc="upper left")
ax.set_ylabel("Defect state percentages [%]")
ax.set_xlabel("Datetime")
ax2 = ax.twinx()
ax2.plot(
timesteps["Datetime"],
timesteps["Pmp_norm"],
c="black",
label="Normalized STC $P_{MP}$",
)
ax2.legend(loc="upper right")
ax2.set_ylabel("Normalized STC $P_{MP}$")
ax.set_title(f"Outdoor LETID \n{location.name}")
plt.show()
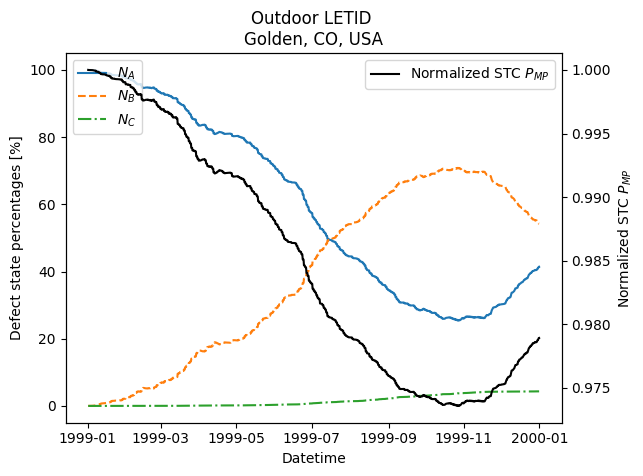
The example data provided for Golden, CO, shows how \(N_A\) increases in cold weather, and power temporarily recovers, due to temporary recovery of LETID (B->A).
The function calc_letid_outdoors wraps all of the steps above into a single function:#
nA_0 = 100
nB_0 = 0
nC_0 = 0
mechanism_params = "repins"
letid.calc_letid_outdoors(
tau_0,
tau_deg,
wafer_thickness,
s_rear,
nA_0,
nB_0,
nC_0,
weather,
meta,
mechanism_params,
generation_df,
module_parameters=cec_module,
)
| Temperature | Injection | NA | NB | NC | tau | Jsc | Voc | Isc | FF | Pmp | Pmp_norm | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | ||||||||||||
| 1999-01-01 00:30:00-07:00 | 0.0 | NaN | 100.000000 | 0.000000e+00 | 0.000000e+00 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| 1999-01-01 01:30:00-07:00 | 0.0 | NaN | 100.000000 | 1.702422e-15 | 0.000000e+00 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| 1999-01-01 02:30:00-07:00 | 0.0 | NaN | 100.000000 | 3.404843e-15 | 5.403329e-36 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| 1999-01-01 03:30:00-07:00 | 0.0 | NaN | 100.000000 | 5.107265e-15 | 1.620999e-35 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| 1999-01-01 04:30:00-07:00 | 0.0 | NaN | 100.000000 | 6.809686e-15 | 3.241997e-35 | 115.000000 | 41.590997 | 0.666327 | 10.106612 | 0.840987 | 5.663467 | 1.000000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1999-12-31 19:30:00-07:00 | 0.0 | NaN | 27.833654 | 6.778463e+01 | 4.381716e+00 | 66.112141 | 41.333851 | 0.654974 | 10.044126 | 0.838966 | 5.519257 | 0.974537 |
| 1999-12-31 20:30:00-07:00 | 0.0 | NaN | 27.833654 | 6.778463e+01 | 4.381716e+00 | 66.112141 | 41.333851 | 0.654974 | 10.044126 | 0.838966 | 5.519257 | 0.974537 |
| 1999-12-31 21:30:00-07:00 | 0.0 | NaN | 27.833654 | 6.778463e+01 | 4.381716e+00 | 66.112141 | 41.333851 | 0.654974 | 10.044126 | 0.838966 | 5.519257 | 0.974537 |
| 1999-12-31 22:30:00-07:00 | 0.0 | NaN | 27.833654 | 6.778463e+01 | 4.381716e+00 | 66.112141 | 41.333851 | 0.654974 | 10.044126 | 0.838966 | 5.519257 | 0.974537 |
| 1999-12-31 23:30:00-07:00 | 0.0 | NaN | 27.833654 | 6.778463e+01 | 4.381716e+00 | 66.112141 | 41.333851 | 0.654974 | 10.044126 | 0.838966 | 5.519257 | 0.974537 |
8760 rows × 12 columns
