Arrhenius Monte Carlo#
A monte carlo simulation can be used to predict results of an event with a certain amount of uncertainty. This will be introduced to our use case via mean and standard deviation for each modeling constant. Correlated multivariate monte carlo simulations expand on this by linking the behavior of multiple input variables together with correlation data, in our case we will use correlation coefficients but
Objectives
Define necessary monte carlo simulation parameters : correlation coefficients, mean and standard standard deviation, number of trials, function to apply, requried function input
Define process for creating and utilizing modeling constant correlation data
Preform simple monte carlo simulation using arrhenius equation to calculate degredation and plot
# if running on google colab, uncomment the next line and execute this cell to install the dependencies and prevent "ModuleNotFoundError" in later cells:
# !pip install pvdeg
import pvlib
import numpy as np
import pandas as pd
import pvdeg
import matplotlib.pyplot as plt
# This information helps with debugging and getting support :)
import sys
import platform
print("Working on a ", platform.system(), platform.release())
print("Python version ", sys.version)
print("Pandas version ", pd.__version__)
print("Pvlib version ", pvlib.__version__)
print("pvdeg version ", pvdeg.__version__)
Working on a Windows 11
Python version 3.13.5 | packaged by Anaconda, Inc. | (main, Jun 12 2025, 16:37:03) [MSC v.1929 64 bit (AMD64)]
Pandas version 2.3.3
Pvlib version 0.13.0
pvdeg version 0.5.1.dev857+g1a511e445.d20251004
Defining Correlation Coefficients#
pvdeg.montecarlo stores correlation coefficients in a Corr object. To represent a given correlation coefficient follow the given syntax below, replacing the values in the brackets with your correlation coefficients
{my_correlation_object} = Corr('{variable1}', '{variable2}', {correlation coefficient})
note: ordering of variable1 and variable2 does not matter
After defining the all known correlations add them to a list which we will feed into our simulation later
corr_Ea_X = pvdeg.montecarlo.Corr("Ea", "X", 0.0269)
corr_Ea_LnR0 = pvdeg.montecarlo.Corr("Ea", "LnR0", -0.9995)
corr_X_LnR0 = pvdeg.montecarlo.Corr("X", "LnR0", -0.0400)
corr_coeff = [corr_Ea_X, corr_Ea_LnR0, corr_X_LnR0]
type(corr_Ea_X)
pvdeg.montecarlo.Corr
Defining Mean and Standard Deviation#
We will store the mean and correlation for each variable, expressed when we defined the correlation cefficients. If a variable is left out at this stage, the monte carlo simulation will throw errors.
stats_dict = {
"Ea": {"mean": 62.08, "stdev": 7.3858},
"LnR0": {"mean": 13.7223084, "stdev": 2.47334772},
"X": {"mean": 0.0341, "stdev": 0.0992757},
}
# and number of monte carlo trials to run
n = 20000
Generating Monte Carlo Input Data#
Next we will use the information collected above to generate correlated data from our modeling constant correlations, means and standard deviations.
np.random.seed(42) # for reproducibility
mc_inputs = pvdeg.montecarlo.generateCorrelatedSamples(
corr=corr_coeff, stats=stats_dict, n=n
)
print(mc_inputs)
Ea LnR0 X
0 65.748631 12.521613 -0.021520
1 61.058808 14.086270 0.113508
2 66.863698 12.047910 0.056494
3 73.328794 10.002535 0.022482
4 60.350590 14.184631 0.159603
... ... ... ...
19995 64.944415 12.851118 -0.074338
19996 72.252954 10.338032 -0.072373
19997 64.874447 12.835188 0.022460
19998 74.735788 9.549980 -0.191887
19999 50.115596 17.754769 0.014540
[20000 rows x 3 columns]
Sanity Check#
We can observe the mean and standard deviation of our newly correlated samples before using them for calculations to ensure that we have not incorrectly altered the data. The mean and standard deviation should be the similar (within a range) to your original input (the error comes from the standard distribution of generated random numbers)
This also applies to the correlation coefficients originally inputted, they should be witin the same range as those orginally supplied.
# mean and standard deviation match inputs
for col in mc_inputs.columns:
print(
f"{col} : mean {mc_inputs[col].mean():.10f}, stdev {mc_inputs[col].std():.10f}"
)
print()
print("Ea_X", round(np.corrcoef(mc_inputs["Ea"], mc_inputs["X"])[0][1], 3))
print("Ea_lnR0", round(np.corrcoef(mc_inputs["Ea"], mc_inputs["LnR0"])[0][1], 3))
print("X_lnR0", round(np.corrcoef(mc_inputs["X"], mc_inputs["LnR0"])[0][1], 3))
Ea : mean 62.1220919316, stdev 7.4023636577
LnR0 : mean 13.7074363705, stdev 2.4773224845
X : mean 0.0349013839, stdev 0.0996405123
Ea_X 0.013
Ea_lnR0 -1.0
X_lnR0 -0.026
Other Function Requirements#
Based on the function chosen to run in the monte carlo simulation, various other data will be required. In this case we will need cell temperature and total plane of array irradiance.
# Load pre-saved weather data for this tutorial
# This avoids API rate limits during testing and builds
import json
weather_df = pd.read_csv("../data/psm4_miami.csv", index_col=0, parse_dates=True)
with open("../data/meta_miami.json", "r") as f:
meta = json.load(f)
# Uncomment below to fetch fresh data with your own API key:
# weather_db = "PSM4"
# weather_id = (25.783388, -80.189029)
# weather_arg = {
# "api_key": "YOUR_API_KEY", # Get your key at https://developer.nrel.gov/signup/
# "email": "your.email@example.com",
# "map_variables": True,
# }
# weather_df, meta = pvdeg.weather.get(weather_db, weather_id, **weather_arg)
Calculate the sun position, poa irradiance, and module temperature.
sol_pos = pvdeg.spectral.solar_position(weather_df, meta)
poa_irradiance = pvdeg.spectral.poa_irradiance(weather_df, meta)
temp_mod = pvdeg.temperature.module(
weather_df=weather_df, meta=meta, poa=poa_irradiance, conf="open_rack_glass_polymer"
)
# the function being used in the monte carlo simulation takes numpy arrays so we need to convert from pd.DataFrame to np.ndarray with .tonumpy()
poa_global = poa_irradiance["poa_global"].to_numpy()
cell_temperature = temp_mod.to_numpy()
The array surface_tilt angle was not provided, therefore the latitude of 25.8 was used.
The array azimuth was not provided, therefore an azimuth of 180.0 was used.
# must already be numpy arrays
function_kwargs = {"poa_global": poa_global, "module_temp": cell_temperature}
Runs monte carlo simulation for the example pvdeg.montecarlo.vecArrhenius function, using the correlated data dataframe created above and the required function arguments.
We can see the necessary inputs by using the help command:
# NBVAL_SKIP
help(pvdeg.montecarlo.simulate)
Help on function simulate in module pvdeg.montecarlo:
simulate(
func: Callable,
correlated_samples: pandas.core.frame.DataFrame,
**function_kwargs
) -> pandas.core.series.Series
Apply a target function to data to preform a monte carlo simulation.
If you get
a key error and the target function has default parameters, try adding them to your
``func_kwargs`` dictionary instead of using the default value from the target
function.
Parameters
----------
func : function
Function to apply for monte carlo simulation
correlated_samples : pd.DataFrame
Dataframe of correlated samples with named columns for each appropriate modeling
constant, can be generated using generateCorrelatedSamples()
function_kwargs : dict
Keyword arguments to pass to func, only include arguments not named in your
correlated_samples columns
Returns
-------
res : pandas.Series
Series with monte carlo results from target function
Running the Monte Carlo Simulation#
We will pass the target function, pvdeg.degredation.vecArrhenius(), its required arguments via the correlated_samples and func_kwargs. Our fixed arguments will be passed in the form of a dictionary while the randomized monte carlo input data will be contained in a DataFrame.
All required target function arguments should be contained between the column names of the randomized input data and fixed argument dictionary,
(You can use any data you want here as long as the DataFrame’s column names match the required target function’s parameter names NOT included in the kwargs)
results = pvdeg.montecarlo.simulate(
func=pvdeg.degradation.vecArrhenius, correlated_samples=mc_inputs, **function_kwargs
)
Viewing Our Data#
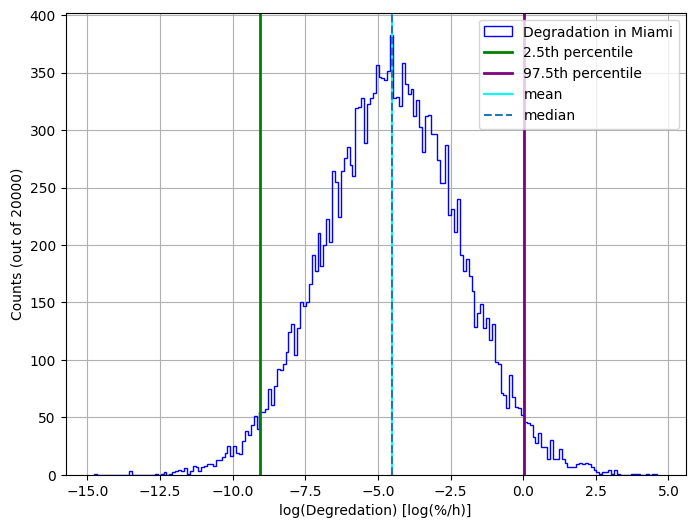
Let’s plot the results using a histogram
lnDeg = np.log10(results)
percentile_2p5 = np.percentile(lnDeg, 2.5)
percentile_97p5 = np.percentile(lnDeg, 97.5)
bin_edges = np.arange(lnDeg.min(), lnDeg.max() + 0.1, 0.1)
plt.figure(figsize=(8, 6))
plt.hist(
lnDeg,
bins=bin_edges,
edgecolor="blue",
histtype="step",
linewidth=1,
label="Degradation in Miami",
)
plt.axvline(percentile_2p5, color="green", label="2.5th percentile", linewidth=2.0)
plt.axvline(percentile_97p5, color="purple", label="97.5th percentile", linewidth=2.0)
plt.axvline(np.mean(lnDeg), color="cyan", label="mean")
plt.axvline(np.median(lnDeg), linestyle="--", label="median")
plt.xlabel("log(Degredation) [log(%/h)]")
plt.ylabel(f"Counts (out of {n})")
plt.legend()
plt.grid(True)
plt.show()